Pooled Screening overview
In pooled shRNA screening, hundreds to thousands of hairpins are combined (pooled) and interrogated simultaneously in a multiplex assay without the need for robotics or liquid handling.
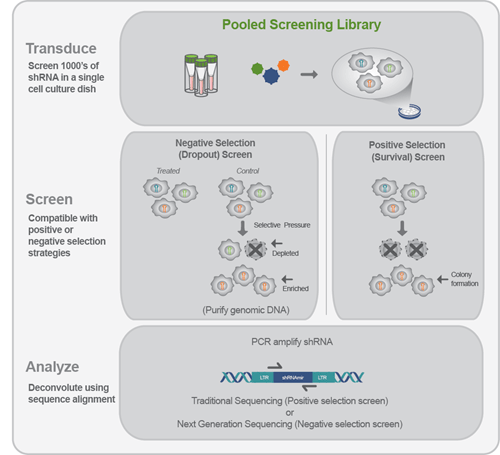
The schematic to the below depicts this process.
- 1. Transduction: Cells are transduced at a low MOI ensuring a single shRNA is expressed in each cell.
- 2. Screen: The screen employs a selection process that is specific to the researcher’s assay. There are two underlying strategies for selection: Negative selection (dropout) screens include an untreated control for comparison to allow the detection of shRNA that provided resistance or sensitized the cells to the selective reagent. Positive selection (enrichment) screens only detect surviving cells and do not require an untreated control.
- 3. Analysis: Under selection, resistant cells increase in the population and sensitized cells decrease in the population. These changes in representation can be detected by sequence analysis (either Sanger sequencing or Next Generation Sequencing).
Pooled screening assays
The underlying workflow of all pooled shRNA screens begins by transducing cells with a heterogeneous pool of viral particles followed by an assay that enriches specific cells based on a phenotypic change. Many variations have been successfully used in vivoor in vitro. However, most fall into three categories: viability screen, reporter screen or behavioral screen1,2
Viability Screen:
In viability screens, the pool is most often used in combination with a selective agent (e.g. drug treatment, exposure to pathogens). An shRNA’s impact on cell health is measured by changes in its representation in population. A negative impact on cell viability will decrease an shRNA’s representation while a positive impact on cell viability will increase representation3,4,5.
Reporter Screens:
Reporters, such as fluorescent markers, can be used to indicate changes in transcription or can be fused to proteins to measure stability or localization. Cells can be sorted for high or low levels of expression and hairpins from each group can be analyzed for their enrichment or depletion8.
Behavioral Screens:
Cell behavior can also be assessed in a pooled screening assay. For example, colony formation on soft agar can be used to select for cells with anchorage independent growth which can be an indicator of oncogenic transformation9,10.
In addition, one of the advantages of shRNA over siRNA is the ability to perform these screens in vivo as well as in vitro. Viability screens have been successfully adapted using xenographs to identify tumor suppressor genes as well as oncogenes5,6,7.
This range of application in addition to the accessibility of shRNA pools provides researchers with a powerful option for screening.
References
1. Sims D. et al. (2011) High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome Biology, 12(10):R104
2. Hu G and Luo J. (2012) A primer on using pooled shRNA libraries for functional genomic screens. Acta Biochim Biophys Sin. Feb;44(2):103-12
3. Westerman B. et al. (2011) A genome-wide RNAi screen in mouse embryonic stem cells identifies Mp1 as a key mediator of differentiation. J Exp Med. Dec 19; 208(13): 2675–2689
4. Schlabach M. et al. (2008) Cancer Proliferation Gene Discovery Through Functional Genomics Science 1 February 2008: Vol. 319 no. 5863 pp. 620-624
5. Luo, J. et al. (2009) A Genome-wide RNAi Screen Identifies Multiple Synthetic Lethal Interactions with the Ras Oncogene. Cell Volume 137, Issue 5, p835–848, 29 May
6. Wuestefeld, T. et al. (2013) A Direct In Vivo RNAi Screen Identifies MKK4 as a Key Regulator of Liver Regeneration. Cell Apr 11; 153(2): 389–401
7. Possemato, R. et al. (2011) Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature Aug 18; 476(7360): 346–350
8. Zender, L. et al. (2008) An Oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. Nov 28; 135(5): 852–864
9. Gazin , C. et al. (2007) An Elaborate Pathway Required for Ras-Mediated Epigenetic Silencing. Nature, Oct 25; 449(7165): 1073-1077
10. Smolen G. et al. (2010) A genome-wide RNAi screen identifies multiple RSK-dependent regulators of cell migration. Genes Dev ;24:v2654-2665.
11. Westbrook T. et al. (2005) A genetic screen for candidate tumor suppressors identifies REST. Cell. 121(6): 837–848.