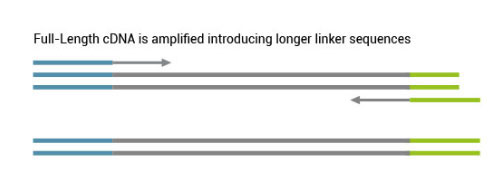
1. How long does the generation of full-length double-stranded DNA take?
The full-length cDNA synthesis takes around 300 minutes in total, including just 60 minutes of hands-on time. For the full-length cDNA amplification using the qPCR and the endpoint PCRs another 300 minutes have to be considered, however only 25 minutes are hands-on time.
2. What are the input RNA requirements?
The protocol was extensively tested with input amounts of 1 ng to 2 µg of total RNA input using RNA extracted from mouse liver, kidney, brain, lung, spleen, thymus, and heart as well as Universal Human Reference RNA and Human Brain Reference RNA.Please consult
Appendix A: RNA Requirements (page 18), TeloPrime User Guide for more details.
3. It is recommended in the User Guide to perform qPCR. What is the reason for this? Can this step be skipped?
Performing a qPCR assay is recommended to determine the exact number of cycles for your endpoint PCR, thereby preventing under- or overcycling which could have negative effects on downstream applications. While you won´t have enough material when undercycling, overcycling will result in a loss of long full-length ds products. If the number of cycles for the endpoint PCR is once established for your specific RNA, it can be used for the following experiments as well without prior qPCR.
4. How is the cycle number of the endpoint PCR determined?
You can determine the cycle number of the endpoint PCR in a qPCR assay by addition of SYBR Green I (final concentration of 0.1x) to the mastermix and using 40 cycles to reach the plateau. Calculate where the fluorescence is at 80% of the maximum; this is the cycle number to be used for the endpoint PCR.Please consult
Appendix B: Calculation of the Endpoint PCR (page 20), TeloPrime User Guide for more details.
5. How many PCR reactions can I do using the TeloPrime kit?
The TeloPrime kit offers you enough reagents to perform one qPCR and one endpoint PCR for each sample. Lexogen also offers a TeloPrime PCR Add-on Kit (Cat.No. 018.30) with additional 30 reactions if you want to use the sample for more PCR reactions.
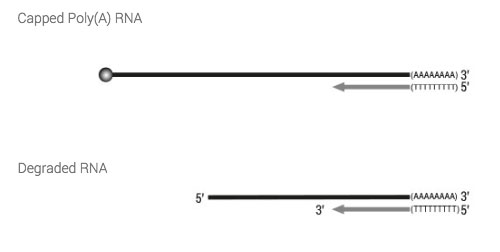
6. How is the 3’ end tagged?
Tagging of the 3’ end of the mRNA takes place during the reverse transcription by oligodT priming of the total RNA.
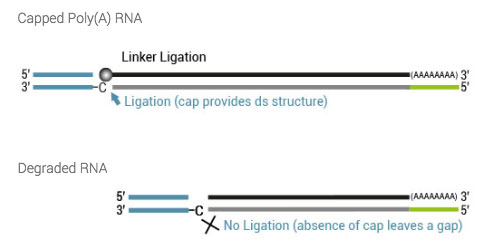
7. How is the 5’ end tagged?
Tagging on the 5’ end is based on Lexogen´s proprietary Cap Dependent Linker Ligation (CDLL) technology. After reverse transcription and the creation of a double-stranded RNA : cDNA hybrid an adapter with a 5’ C overhang allows for an atypical base-pairing with the inverted G of the cap structure. Hence, ligation is taking place using a double-strand specific ligase by ligating the 3’ end of the linker to the 3’ end of the cDNA.
8. Which sequences are used for PCR amplification?
The PCR primers included in the TeloPrime Kit have the following sequence.
FP: 5’ – TGGATTGATATGTAATACGACTCACTATAG – 3‘ RP: 5’ – TCTCAGGCGTTTTTTTTTTTTTTTTTT – 3‘
The FP contains the T7 promoter sequence.
9. Can I use my own primers instead of Reverse Transcription, PCR Forward, and PCR Reverse Primer?
The Reverse Transcription, PCR Forward, and PCR Reverse Primer are added separately in theTeloPrime protocol and can easily be exchanged. If exchanging the RT-primer to another oligodT or gene-specific primer just make sure to use the same sequence in the PCR amplification afterwards. Please keep in mind, the adapter sequences cannot be exchanged.
10. What downstream applications can the full-length cDNA be used for?
The full-length cDNA products can be used for Next Generation Sequencing (NGS) library preparation, RACE, cloning, microarray probes, and normalization.
11. Which NGS library preparation kit can be used after generation of cDNA with TeloPrime?
Any commercially available DNA-Seq library prep kit can be used on top of the TeloPrime kit in order to produce sequence-ready libraries.
12. Is TeloPrime compatible with the SENSE Total RNA/mRNA-Seq Library Prep Kits?
No, the TeloPrime kits generate full-length double-stranded DNA and the SENSE kits are RNA-Seq library prep kits and need RNA as an input material.